Summer 2015
Advanced molecular detection promises advances in both laboratory and epidemiological practice, especially in the area of whole genome sequencing (WGS). In the spring of 2015, APHL and CDC initiated a pilot project, in collaboration with the Wisconsin State Laboratory of Hygiene (WSLH), to exchange WGS data between APHL Influenza Reference Centers and CDC. The result is an elegantly simple deployment of technological tools that, when integrated, create an accessible environment for data exchange, management and analysis of WGS data.
13 GB and Counting
But this solution was not easily achieved. From the outset, the challenges of a new technology that generates a 13 GB file with a single full run were evident: data storage, inadequate broadband speeds and infrastructure, CDC firewall restrictions, expensive bioinformatics tools and long -term archiving support. To address these issues, APHL worked closely with the WSLH and CDC’s Influenza Division to explore and pilot WGS data exchange options between the Influenza Reference Centers and CDC.
The multi-faceted solution relies on the APHL Informatics Messaging Services (AIMS) environment. Developed by APHL with CDC support, the AIMS platform was designed to enable public health entities to share data efficiently and securely with diverse messaging partners. Since then, it has evolved into a stable and secure environment whose application and platform services reside in the Amazon Web Services cloud environment.
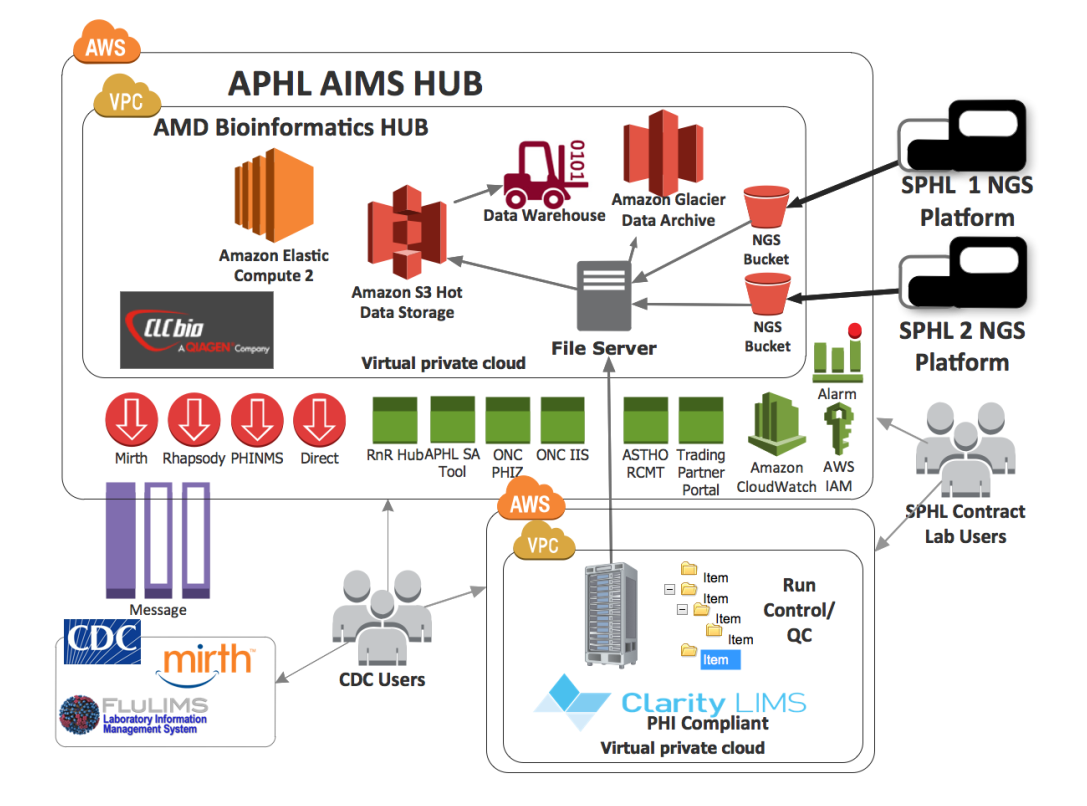 The solution begins with the transfer of Wisconsin WGS data using AWS Simple Storage Service (S3) client -- a tool that simplifies the uploading and downloading of WGS data to AIMS via a client-side application. Once the data is secured on AIMS, raw sequence files are available to CDC. These files will ultimately be available to submitting reference center labs through a simple file management service.
The solution begins with the transfer of Wisconsin WGS data using AWS Simple Storage Service (S3) client -- a tool that simplifies the uploading and downloading of WGS data to AIMS via a client-side application. Once the data is secured on AIMS, raw sequence files are available to CDC. These files will ultimately be available to submitting reference center labs through a simple file management service.
WGS raw data can then be transferred to bioinformatics tools for data analysis and to a separately managed/secured AWS cloud-based instance of Clarity LIMS to support quality control and run management. After two to three weeks, data can be transferred to AWS Glacier for long-term archiving and online backup.
Promising Results
The pilot project has shown very promising results. Large, multi-gigabit data files are available to trading partners in only a few minutes, a process that often took several days previously. Analysis is faster and analytical results, notes and updates are shared among all participants simultaneously from a central data source. Data is secured in a centralized environment using processes and procedures that comply with multiple federal standards.
The APHL/CDC Whole Genome Sequencing Data Exchange project holds great promise for innovation, data exchange, data availability and analytics. As it advances over the next year, APHL and CDC will add other reference centers and laboratories with the goal of making the system available to all public health laboratories. In addition, the partners plan to expand the scope beyond influenza to include several other pathogens.
For more information, contact Patina Zarcone, director, Informatics, 240.485.2788, patina.zarcone@aphl.org